XML Variant Ingestion
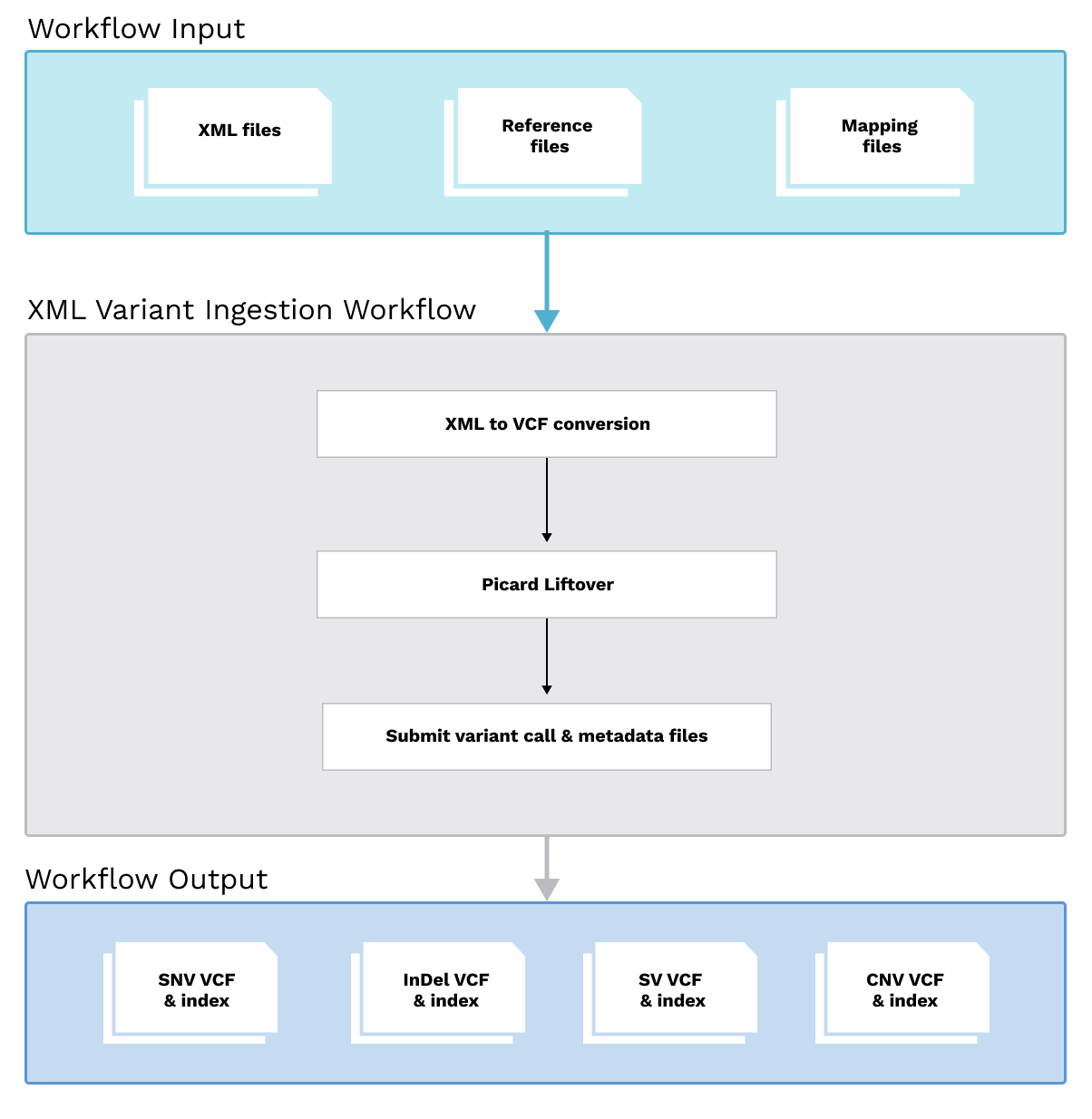
The ARGO Data Platform supports the ingestion of variant calling data submitted in XML format (based on the GRCh37/hg19 genome reference). These XML files are processed through a standardized workflow to convert them into VCF format, followed by a liftover to the GRCh38 reference genome. For details, please see the latest version of the ARGO xml_variant_ingestion workflow.
Inputs
- Submitted XML file(s): Containing variant calls based on the GRCh37 (hg19) genome reference.
- Metadata mapping file: Used to map identifiers and provide additional sequencing and variant calling context.
- Human reference genome:
- Genome liftover chain file: Required for coordinate transformation from GRCh37 to GRCh38.
Processing
XML to VCF Conversion:
- Variant records are parsed from XML and converted into corresponding VCF records.
- Variants are separated by type:
- Short Variants: SNVs and InDels
- Copy Number Alterations (CNVs)
- Structural Variants / Rearrangements (SVs)
Reference Genome Liftover:
- The converted VCF files are lifted from GRCh37 to GRCh38 using Picard:liftovervcf.
Outputs
- Raw SNV Calls and VCF Index
- Raw Indel Calls and VCF Index
- Raw SV Calls and VCF Index
- Raw CNV Calls and VCF Index
Workflow Diagram