Analysis Pipelines Overview
Molecular data submitted to the ARGO Data Platform will undergo molecular analysis through a unified pipeline. Consistent pipeline analysis across different samples ensures that data can be considered functionally equivalent across different data sets, including all ARGO programs as well as any other initiatives that have adopted the ARGO analysis pipelines, increasing the research power of the dataset provided by the ARGO Data Platform.
The ARGO Data Platform will accept a wide range of datatypes, including:
- Genomic data from both Whole Exome Sequencing (WXS) and Whole Genome Sequencing (WGS)
- DNA Methylation data
- Transcriptomic data
- Proteomic data
- Variant calling data (in XML format)
- Slide images
Pipelines, and the individual analysis workflows that they are constructed from, have been developed by the DCC Bioinformatics team based on established, best community practices.
All ARGO analysis workflows are written in Nextflow. Nextflow is developed by the Comparative Bioinformatics group at the Barcelona Centre for Genomic Regulation (CRG).
All ARGO workflows are published under an GNU AGPL v3 open-source license and are packaged for community usage.
DNA-Sequencing Analysis Pipeline
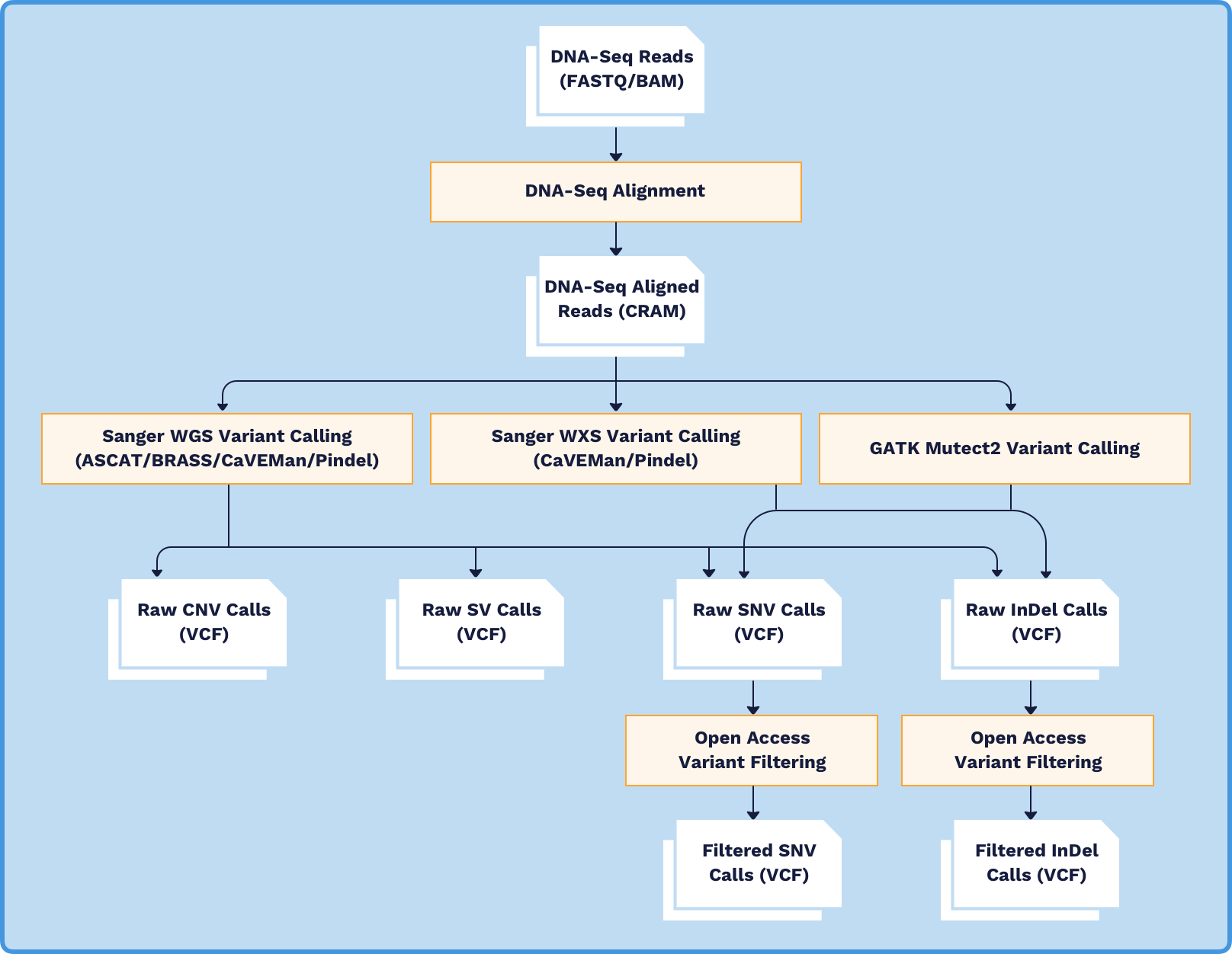
The DNA-Sequencing (DNA-Seq) analysis pipeline identifies multiple types of somatic variant from both Whole Exome Sequencing (WXS) and Whole Genome Sequencing (WGS) sample data. DNA-Seq analysis consists of the following workflows:
- DNA-Seq Alignment
- Sanger WGS Variant Calling
- Sanger WXS Variant Calling
- GATK Mutect2 Variant Calling
- Open Access Variant Filtering
In the future, these procedures will be extended to include:
- Somatic Variant Calling
- Germline Variant Calling
- Consensus Variant Calling
- Variant Masking
- Variant Annotation
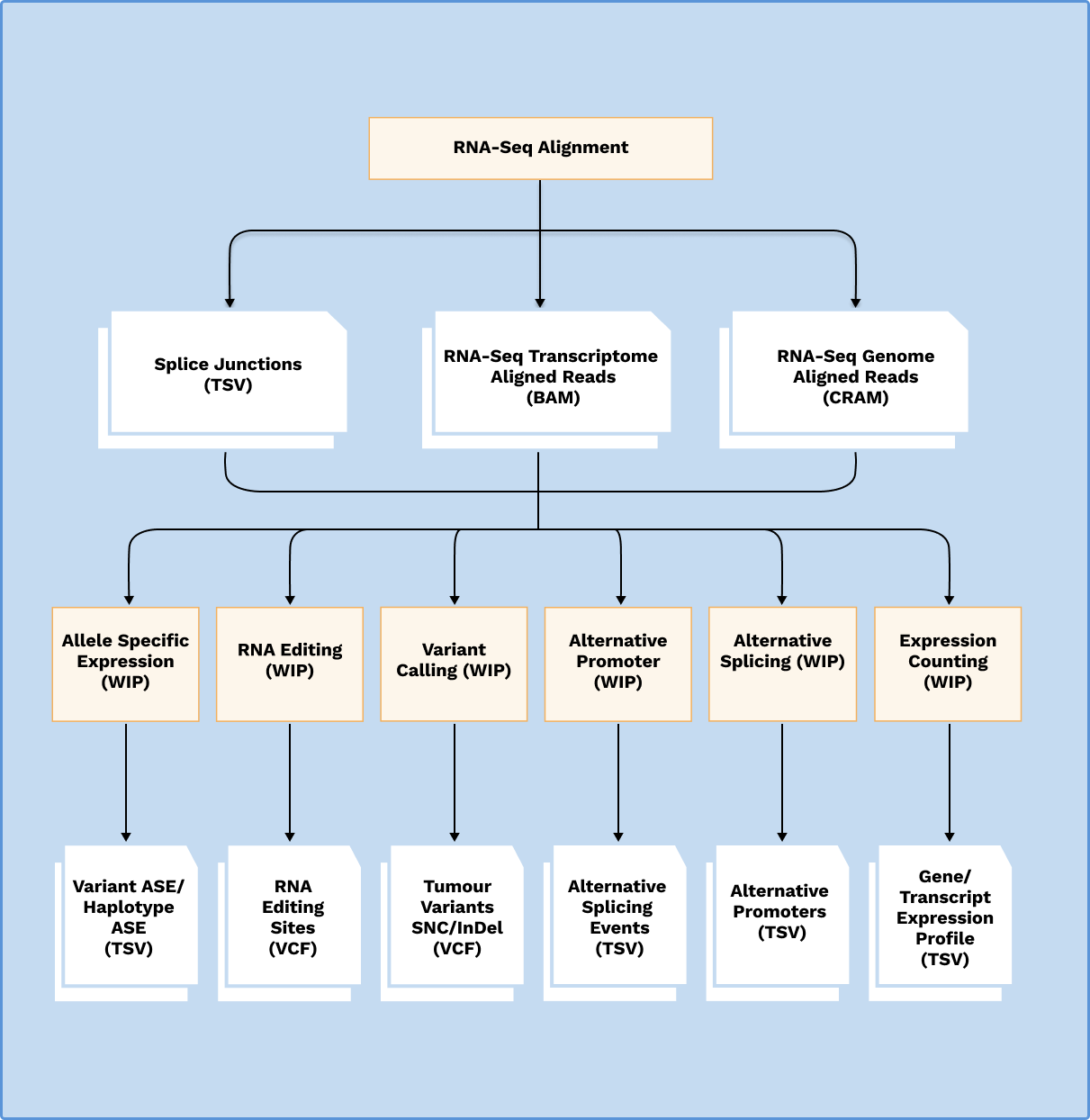
RNA-Sequencing Analysis Pipeline
The RNA-Sequencing (RNA-Seq) analysis pipeline identifies gene expression patterns and differentially expressed genes between samples or conditions, and to gain insights into their biological functions and pathways. RNA-Seq analysis consists and will be extended to include:
- RNA-Seq Alignment
- Expression Counting
- Alternative Promoter
- Alternative Splicing
- RNA Variant Calling
- Allele Specific Expression
- RNA Editing
- RNA Fusion